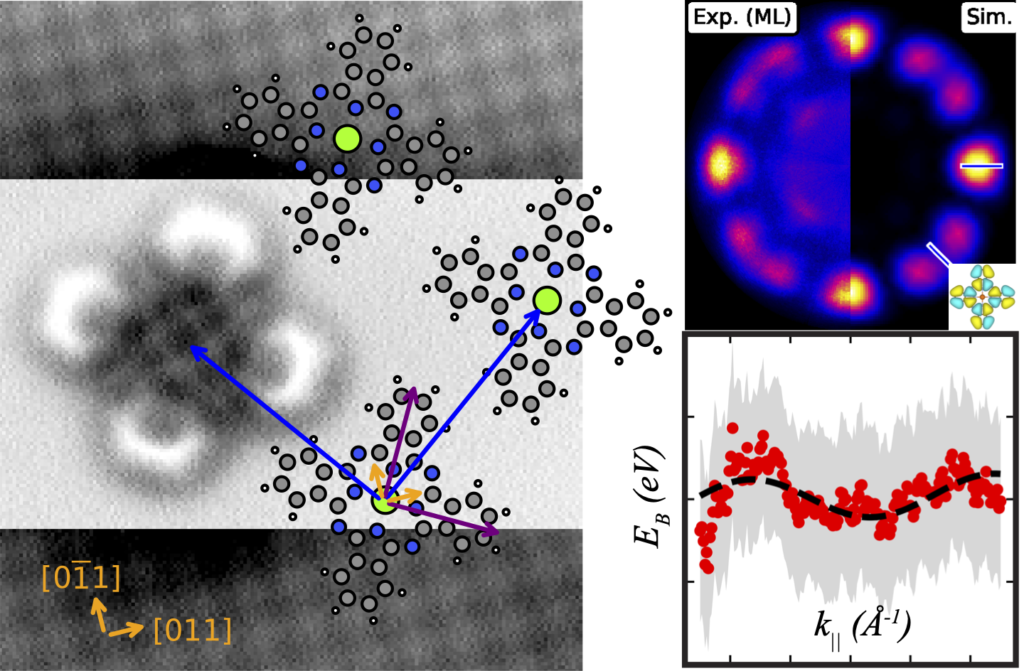
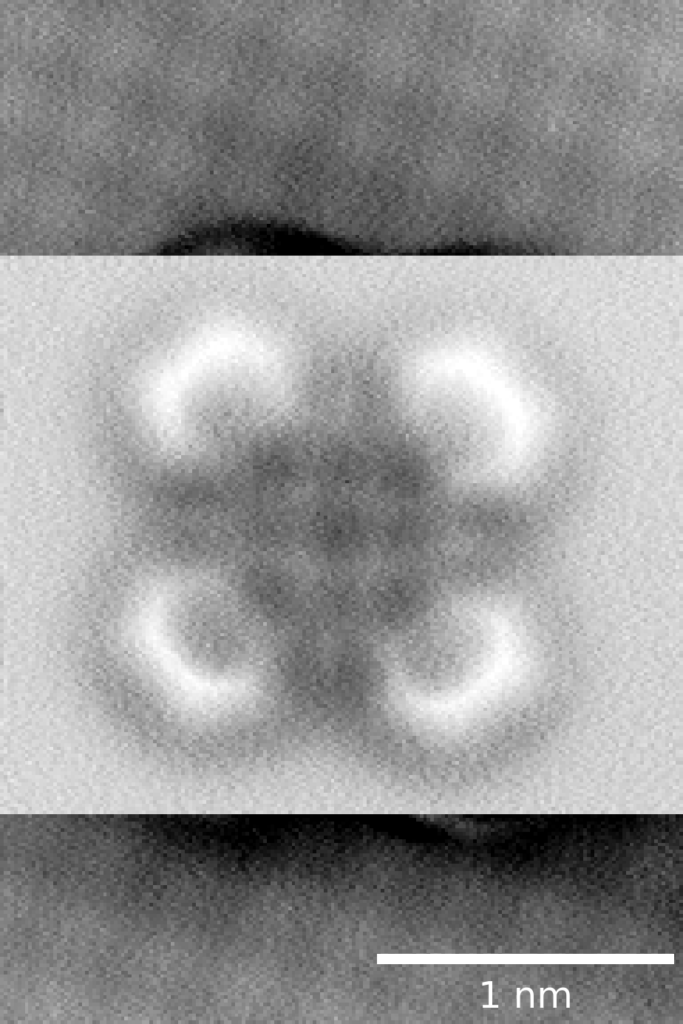
Jules has put in the hard yards implementing nanonisTCP as a python module, and leveraged that to create scanbot, a tool for automating the tasks of preparing a good imaging & spectroscopy probe, as well as a suite of functions for performing nuanced, drift-corrected measurements over very long timescales.
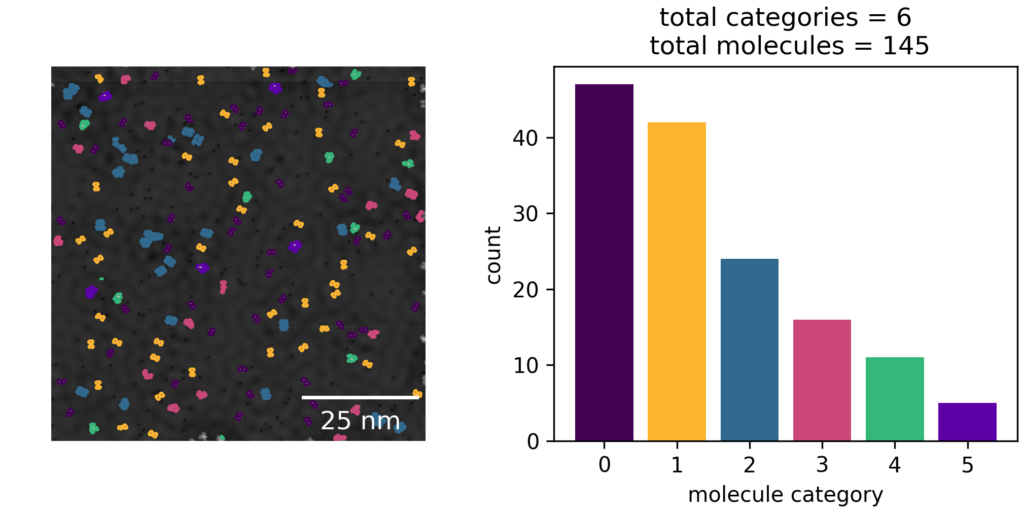
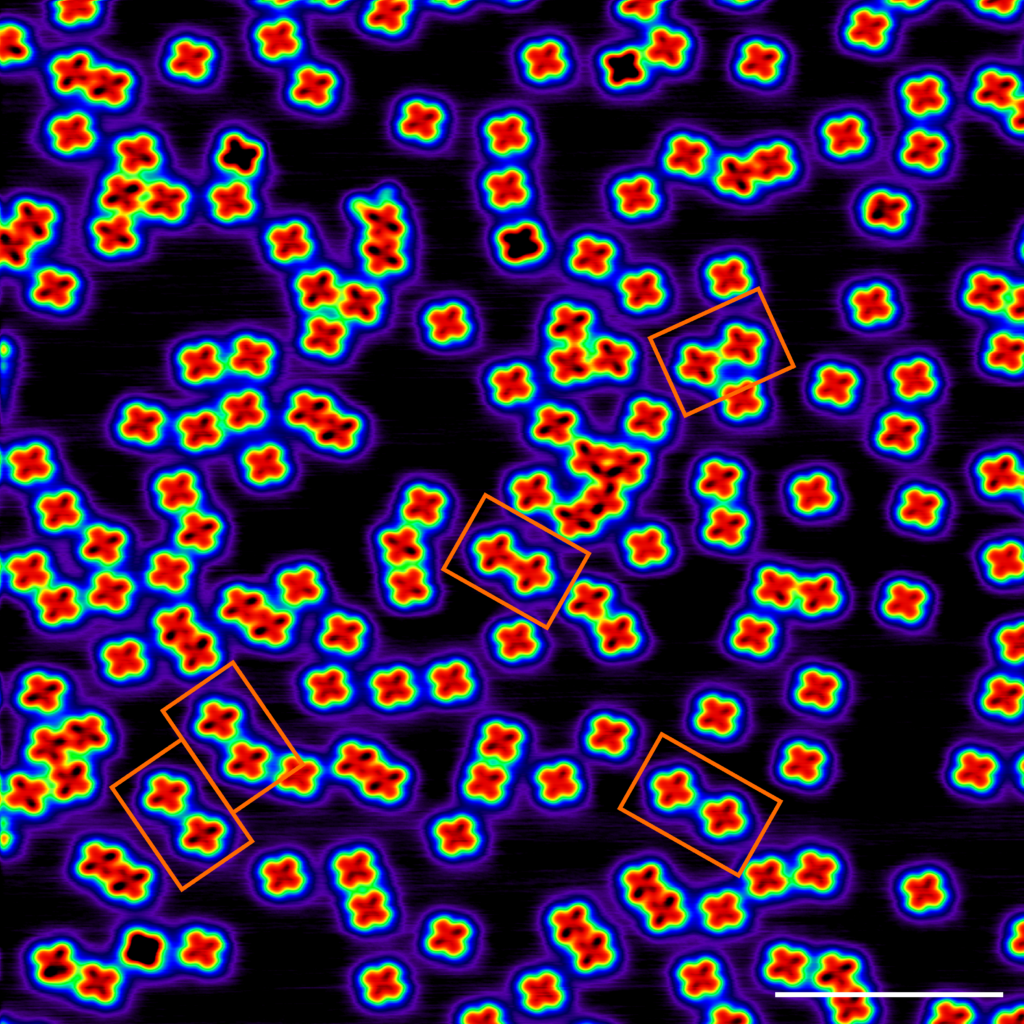
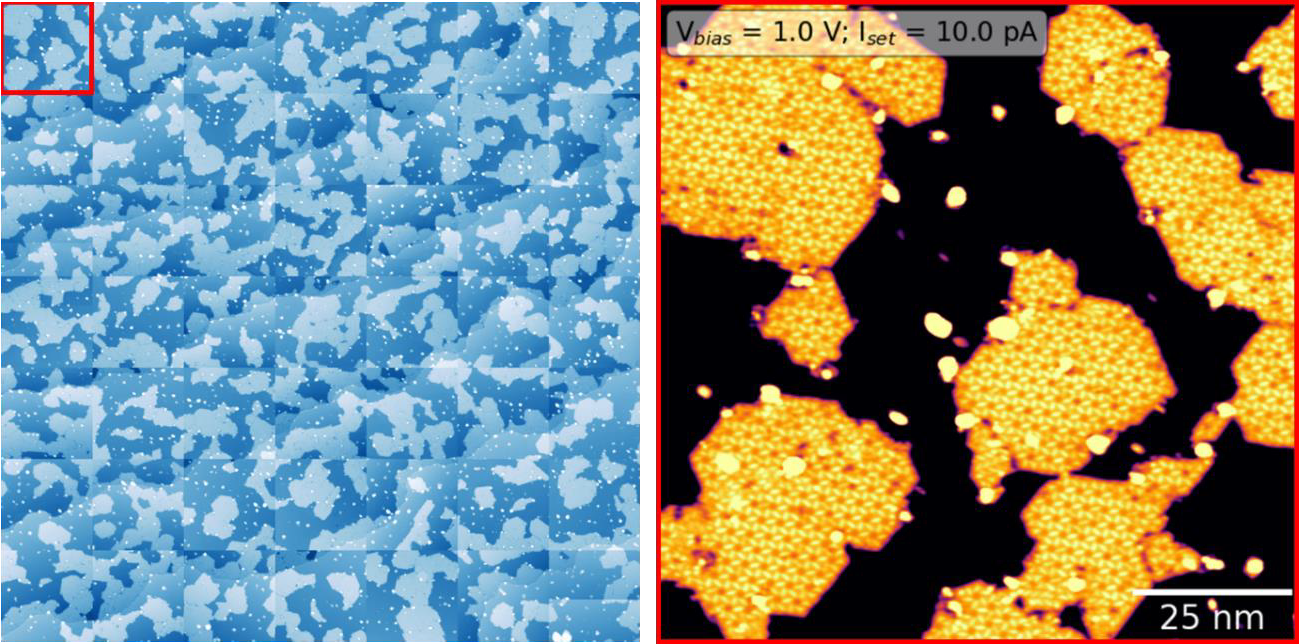
See the below example of systematically grid scanning 100x100nm images to concatenate a comprehensive view of the surface. Right is the upper left red corner, where self-assembled molecular islands are visible.
Ceddia et al., (2024). Scanbot: An STM Automation Bot. Journal of Open Source Software, 9(99), 6028, https://doi.org/10.21105/joss.06028